echopype Tour
Contents
1. echopype Tour#
https://github.com/OSOceanAcoustics/echopype-examples/blob/main/notebooks/
A quick tour of core echopype capabilities.
1.1. Introduction#
1.1.1. Goals#
Illustrate a common workflow for echosounder data conversion, calibration and use. This workflow leverages the standardization applied by echopype and the power, ease of use and familiarity of libraries in the scientific Python ecosystem.
Extract and visualize data with relative ease.
1.1.2. Description#
This notebook uses EK60 echosounder data collected during the 2017 Joint U.S.-Canada Integrated Ecosystem and Pacific Hake Acoustic Trawl Survey (‘Pacific Hake Survey’) to illustrate a common workflow for data conversion, calibration and analysis using echopype and core scientific Python software packages, particularly xarray, GeoPandas and pandas.
Two days of cloud-hosted .raw data files are accessed by echopype directly from an Amazon Web Services (AWS) S3 “bucket” maintained by the NOAA NCEI Water-Column Sonar Data Archive. With echopype, each file is converted to a standardized representation based on the SONAR-netCDF4 v1.0 convention and saved to the Zarr cloud-optimized file format.
Data stored in the Zarr (or netCDF) file format using the SONAR-netCDF4 convention can be conveniently and intuitively manipulated with xarray in combination with related scientific Python packages. Mean Volume Backscattering Strength (MVBS) is computed with echopype from the combined, converted raw data files, after calibration. The GPS location track and MVBS echograms are visualized.
1.1.3. Running the notebook#
This notebook can be run with a conda environment created using the conda environment file https://github.com/OSOceanAcoustics/echopype-examples/blob/main/binder/environment.yml. The notebook creates the ./exports directory, if not already present. Zarr files will be exported there.
from pathlib import Path
import fsspec
import geopandas as gpd
import xarray as xr
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.io.img_tiles as cimgt
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
import hvplot.xarray
import echopype as ep
import warnings
warnings.simplefilter("ignore", category=DeprecationWarning)
1.2. Compile list of raw files to read from the NCEI WCSD S3 bucket#
We’ll compile a list of several EK60 .raw files from the 2017 Pacific Hake survey, available via open, anonymous access from an AWS S3 bucket managed by the NCEI WCSD.
After using fsspec to establish an S3 “file system” access point, extract a list of .raw files in the target directory.
fs = fsspec.filesystem('s3', anon=True)
bucket = "ncei-wcsd-archive"
rawdirpath = "data/raw/Bell_M._Shimada/SH1707/EK60"
s3rawfiles = fs.glob(f"{bucket}/{rawdirpath}/*.raw")
print(f"There are {len(s3rawfiles)} raw files in the directory")
There are 4343 raw files in the directory
# print out the last two S3 raw file paths in the list
s3rawfiles[-2:]
['ncei-wcsd-archive/data/raw/Bell_M._Shimada/SH1707/EK60/Summer2017-D20170913-T180733.raw',
'ncei-wcsd-archive/data/raw/Bell_M._Shimada/SH1707/EK60/Winter2017-D20170615-T002629.raw']
Each of these .raw files is typically about 25 MB. To select a reasonably small but meaningful target for this demo, let’s select all files from 2017-07-28 collected over a two hour period, 18:00 to 19:59 UTC. This is done through string matching on the time stamps found in the file names.
s3rawfiles = [
s3path for s3path in s3rawfiles
if any([f"D2017{dtstr}" in s3path for dtstr in ['0728-T18', '0728-T19']])
]
print(f"There are {len(s3rawfiles)} target raw files available")
There are 5 target raw files available
s3rawfiles
['ncei-wcsd-archive/data/raw/Bell_M._Shimada/SH1707/EK60/Summer2017-D20170728-T181619.raw',
'ncei-wcsd-archive/data/raw/Bell_M._Shimada/SH1707/EK60/Summer2017-D20170728-T184131.raw',
'ncei-wcsd-archive/data/raw/Bell_M._Shimada/SH1707/EK60/Summer2017-D20170728-T190728.raw',
'ncei-wcsd-archive/data/raw/Bell_M._Shimada/SH1707/EK60/Summer2017-D20170728-T193459.raw',
'ncei-wcsd-archive/data/raw/Bell_M._Shimada/SH1707/EK60/Summer2017-D20170728-T195219.raw']
1.3. Read target raw files directly from AWS S3 bucket and convert to netcdf#
Create the directory where the exported files will be saved, if this directory doesn’t already exist.
base_dpath = Path('./exports/notebook1')
base_dpath.mkdir(exist_ok=True, parents=True)
EchoData is an echopype object for conveniently handling raw converted data from either raw instrument files or previously converted and standardized raw netCDF4 and Zarr files. It is essentially a container for multiple xarray.Dataset objects, each corresponds to one of the netCDF4 groups specified in the SONAR-netCDF4 convention – the convention followed by echopype. The EchoData object can be used to conveniently accesse and explore the echosounder raw data and for calibration and other processing.
For each raw file:
Access file directly from S3 via
ep.open_rawto create a convertedEchoDataobject in memoryAdd global and platform attributes to
EchoDataobjectExport to a local Zarr file
for s3rawfpath in s3rawfiles:
raw_fpath = Path(s3rawfpath)
try:
# Access file directly from S3 to create a converted EchoData object in memory
ed = ep.open_raw(
f"s3://{s3rawfpath}",
sonar_model='EK60',
storage_options={'anon': True}
)
# Manually populate additional metadata about the dataset and the platform
# -- SONAR-netCDF4 Top-level Group attributes
ed['Top-level'].attrs['title'] = "2017 Pacific Hake Acoustic Trawl Survey"
ed['Top-level'].attrs['summary'] = (
f"EK60 raw file from the {ed['Top-level'].attrs['title']}, "
"converted to a SONAR-netCDF4 file using echopype."
)
# -- SONAR-netCDF4 Platform Group attributes
ed['Platform'].attrs['platform_type'] = "Research vessel"
ed['Platform'].attrs['platform_name'] = "Bell M. Shimada" # A NOAA ship
ed['Platform'].attrs['platform_code_ICES'] = "315"
# Save to converted Zarr format
# Use the same base file name as the raw file but with a ".zarr" extension
ed.to_zarr(save_path=base_dpath, overwrite=True)
except Exception as e:
print(f"Failed to process raw file {raw_fpath.name}: {e}")
Let’s examine the last EchoData object created above, ed. This summary shows a collapsed view of the netCDF4 groups that make up the EchoData object, where each group corresponds to an xarray Dataset. The backscatter data is in the Sonar/Beam_group1 group, accessible as ed['Sonar/Beam_group1'].
ed
-
<xarray.Dataset> Size: 0B Dimensions: () Data variables: *empty* Attributes: conventions: CF-1.7, SONAR-netCDF4-1.0, ACDD-1.3 keywords: EK60 sonar_convention_authority: ICES sonar_convention_name: SONAR-netCDF4 sonar_convention_version: 1.0 summary: EK60 raw file from the 2017 Pacific Hake Aco... title: 2017 Pacific Hake Acoustic Trawl Survey date_created: 2017-07-28T19:52:19Z processing_level: Level 1A processing_level_url: https://echopype.readthedocs.io/en/stable/pr... -
<xarray.Dataset> Size: 30kB Dimensions: (channel: 3, time1: 523) Coordinates: * channel (channel) <U37 444B 'GPT 18 kHz 009072058c8d 1-1... * time1 (time1) datetime64[ns] 4kB 2017-07-28T19:52:19.12... Data variables: absorption_indicative (channel, time1) float64 13kB 0.002822 ... 0.03259 sound_speed_indicative (channel, time1) float64 13kB 1.481e+03 ... 1.481... frequency_nominal (channel) float64 24B 1.8e+04 3.8e+04 1.2e+05 -
<xarray.Dataset> Size: 117kB Dimensions: (time1: 2769, time2: 523, channel: 3) Coordinates: * time1 (time1) datetime64[ns] 22kB 2017-07-28T19:52:22.0016... * time2 (time2) datetime64[ns] 4kB 2017-07-28T19:52:19.12031... * channel (channel) <U37 444B 'GPT 18 kHz 009072058c8d 1-1 ES... Data variables: (12/20) latitude (time1) float64 22kB 43.69 43.69 43.69 ... 43.78 43.78 longitude (time1) float64 22kB -125.2 -125.2 ... -125.2 -125.2 sentence_type (time1) <U3 33kB 'GGA' 'GLL' 'GGA' ... 'GLL' 'GGA' pitch (time2) float64 4kB -0.4641 -0.2911 ... -1.072 -0.3782 roll (time2) float64 4kB 0.58 -0.4938 ... 0.6955 -0.03356 vertical_offset (time2) float64 4kB -0.1959 -0.00136 ... 0.16 -0.1182 ... ... position_offset_y float64 8B nan position_offset_z float64 8B nan transducer_offset_x (channel) float64 24B 0.0 0.0 0.0 transducer_offset_y (channel) float64 24B 0.0 0.0 0.0 transducer_offset_z (channel) float64 24B 0.0 0.0 0.0 frequency_nominal (channel) float64 24B 1.8e+04 3.8e+04 1.2e+05 Attributes: platform_name: Bell M. Shimada platform_type: Research vessel platform_code_ICES: 315 -
<xarray.Dataset> Size: 9MB Dimensions: (time1: 29194) Coordinates: * time1 (time1) datetime64[ns] 234kB 2017-07-28T19:52:19.120310 ..... Data variables: NMEA_datagram (time1) <U73 9MB '$SDVLW,5063.975,N,5063.975,N' ... '$INHD... Attributes: description: All NMEA sensor datagrams -
<xarray.Dataset> Size: 376B Dimensions: (filenames: 1) Coordinates: * filenames (filenames) int64 8B 0 Data variables: source_filenames (filenames) <U92 368B 's3://ncei-wcsd-archive/data/raw/... Attributes: conversion_software_name: echopype conversion_software_version: 0.8.4 conversion_time: 2024-04-25T21:30:45Z -
<xarray.Dataset> Size: 568B Dimensions: (beam_group: 1) Coordinates: * beam_group (beam_group) <U11 44B 'Beam_group1' Data variables: beam_group_descr (beam_group) <U131 524B 'contains backscatter power (un... Attributes: sonar_manufacturer: Simrad sonar_model: EK60 sonar_serial_number: sonar_software_name: ER60 sonar_software_version: 2.4.3 sonar_type: echosounder -
<xarray.Dataset> Size: 37MB Dimensions: (channel: 3, ping_time: 523, range_sample: 3957) Coordinates: * channel (channel) <U37 444B 'GPT 18 kHz 009072058... * ping_time (ping_time) datetime64[ns] 4kB 2017-07-28T... * range_sample (range_sample) int64 32kB 0 1 2 ... 3955 3956 Data variables: (12/29) frequency_nominal (channel) float64 24B 1.8e+04 3.8e+04 1.2e+05 beam_type (channel) int64 24B 1 1 1 beamwidth_twoway_alongship (channel) float64 24B 10.9 6.81 6.58 beamwidth_twoway_athwartship (channel) float64 24B 10.82 6.85 6.52 beam_direction_x (channel) float64 24B nan nan nan beam_direction_y (channel) float64 24B nan nan nan ... ... data_type (channel, ping_time) int8 2kB 3 3 3 ... 3 3 3 sample_time_offset (channel, ping_time) float64 13kB 0.0 ... 0.0 channel_mode (channel, ping_time) int8 2kB 0 0 0 ... 0 0 0 backscatter_r (channel, ping_time, range_sample) float32 25MB ... angle_athwartship (channel, ping_time, range_sample) int8 6MB ... angle_alongship (channel, ping_time, range_sample) int8 6MB ... Attributes: beam_mode: vertical conversion_equation_t: type_3 -
<xarray.Dataset> Size: 868B Dimensions: (channel: 3, pulse_length_bin: 5) Coordinates: * channel (channel) <U37 444B 'GPT 18 kHz 009072058c8d 1-1 ES18... * pulse_length_bin (pulse_length_bin) int64 40B 0 1 2 3 4 Data variables: frequency_nominal (channel) float64 24B 1.8e+04 3.8e+04 1.2e+05 sa_correction (channel, pulse_length_bin) float64 120B 0.0 ... -0.3 gain_correction (channel, pulse_length_bin) float64 120B 20.3 ... 26.55 pulse_length (channel, pulse_length_bin) float64 120B 0.000512 ... ...
print(f"Number of ping times in this EchoData object: {len(ed['Sonar/Beam_group1'].ping_time)}")
Number of ping times in this EchoData object: 523
Assemble a list of EchoData objects from the converted Zarr files and combine them into a single EchoData object using the combine_echodata function. By default open_converted lazy loads individual Zarr files and only metadata are read into memory, until more operations are executed, which in this case is the combine_echodata function.
ed_list = []
for converted_file in sorted(base_dpath.glob("*.zarr")):
ed_list.append(ep.open_converted(converted_file))
combined_ed = ep.combine_echodata(ed_list)
print(f"Number of ping times in the combined EchoData object: {len(combined_ed['Sonar/Beam_group1'].ping_time)}")
Number of ping times in the combined EchoData object: 2379
The Provenance group now of the combined EchoData object contains some helpful information compiled from the source files.
combined_ed['Provenance']
<xarray.Dataset> Size: 11kB
Dimensions: (filenames: 5, echodata_filename: 5)
Coordinates:
* filenames (filenames) int64 40B 0 1 2 3 4
* echodata_filename (echodata_filename) <U33 660B 'Summer2017-D2...
Data variables: (12/26)
source_filenames (filenames) <U92 2kB 's3://ncei-wcsd-archive...
conventions (echodata_filename) <U35 700B 'CF-1.7, SONAR...
date_created (echodata_filename) <U20 400B '2017-07-28T18...
keywords (echodata_filename) <U4 80B 'EK60' ... 'EK60'
processing_level (echodata_filename) <U8 160B 'Level 1A' ... ...
processing_level_url (echodata_filename) <U64 1kB 'https://echopy...
... ...
sonar_serial_number (echodata_filename) <U1 20B '' '' '' '' ''
sonar_software_name (echodata_filename) <U4 80B 'ER60' ... 'ER60'
sonar_software_version (echodata_filename) <U5 100B '2.4.3' ... '2....
sonar_type (echodata_filename) <U11 220B 'echosounder' ...
beam_mode (echodata_filename) <U8 160B 'vertical' ... ...
conversion_equation_t (echodata_filename) <U6 120B 'type_3' ... 't...
Attributes:
conversion_software_name: echopype
conversion_software_version: 0.8.4
conversion_time: 2024-04-25T21:30:34Z
is_combined: True
combination_software_name: echopype
combination_software_version: 0.8.4
combination_time: 2024-04-25T21:30:45Z1.4. Extract and plot GPS locations from Platform group#
Extract and join the latitude and longitude variables from the Platform group in the combined_ed EchoData object. Convert to a Pandas DataFrame first, then to a GeoPandas GeoDataFrame for convenient viewing and manipulation.
gps_df = combined_ed['Platform'].latitude.to_dataframe().join(combined_ed['Platform'].longitude.to_dataframe())
gps_df.head(3)
| latitude | longitude | |
|---|---|---|
| time1 | ||
| 2017-07-28 18:16:21.476992 | 43.657532 | -124.887015 |
| 2017-07-28 18:16:21.635323 | 43.657500 | -124.887000 |
| 2017-07-28 18:16:22.169931 | 43.657532 | -124.887080 |
gps_gdf = gpd.GeoDataFrame(
gps_df,
geometry=gpd.points_from_xy(gps_df['longitude'], gps_df['latitude']),
crs="epsg:4326"
)
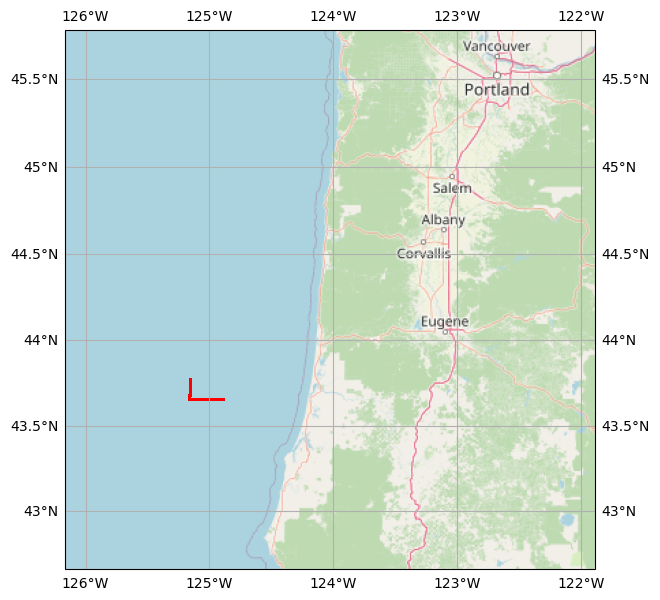
Create a cartopy reference map from the point GeoDataFrame, to place the GPS track in its geographical context.
basemap = cimgt.OSM()
_, ax = plt.subplots(
figsize=(7, 7), subplot_kw={"projection": basemap.crs}
)
bnd = gps_gdf.geometry.bounds
ax.set_extent([bnd.minx.min() - 1, bnd.maxx.max() + 3,
bnd.miny.min() - 1, bnd.maxy.max() + 2])
ax.add_image(basemap, 7)
ax.gridlines(draw_labels=True, xformatter=LONGITUDE_FORMATTER, yformatter=LATITUDE_FORMATTER)
gps_gdf.plot(ax=ax, markersize=0.1, color='red',
transform=ccrs.PlateCarree());
1.5. Compute mean volume backscattering strength (\(\text{MVBS}\)) and plot interactive echograms using hvplot#
echopype supports basic processing funcionalities including calibration (from raw instrument data records to volume backscattering strength, \(S_V\)), denoising, and computing mean volume backscattering strength, \(\overline{S_V}\) or \(\text{MVBS}\). The EchoData object can be passed into various calibration and preprocessing functions without having to write out any intermediate files.
We’ll calibrate the combined backscatter data to obtain \(S_V\). For EK60 data, by default the function uses environmental (sound speed and absorption) and calibration parameters stored in the data file. Users can optionally specify other parameter choices. \(S_V\) is then binned along range (depth) and ping time to generate \(\text{MVBS}\).
Sv_ds = ep.calibrate.compute_Sv(combined_ed).compute()
MVBS_ds = ep.commongrid.compute_MVBS(
Sv_ds,
range_bin= '5m', # in meters
ping_time_bin='20s' # in seconds
)
Sv_ds
<xarray.Dataset> Size: 452MB
Dimensions: (channel: 3, ping_time: 2379,
range_sample: 3957, filenames: 1)
Coordinates:
* channel (channel) <U37 444B 'GPT 18 kHz 009072058...
* ping_time (ping_time) datetime64[ns] 19kB 2017-07-28...
* range_sample (range_sample) int64 32kB 0 1 2 ... 3955 3956
* filenames (filenames) int64 8B 0
Data variables: (12/16)
Sv (channel, ping_time, range_sample) float64 226MB ...
echo_range (channel, ping_time, range_sample) float64 226MB ...
frequency_nominal (channel) float64 24B 1.8e+04 3.8e+04 1.2e+05
sound_speed (channel, ping_time) float64 57kB 1.481e+0...
sound_absorption (channel, ping_time) float64 57kB 0.002822...
sa_correction (channel, ping_time) float64 57kB -0.7 ......
... ...
angle_sensitivity_alongship (channel) float64 24B 13.89 21.97 23.12
angle_sensitivity_athwartship (channel) float64 24B 13.89 21.97 23.12
beamwidth_alongship (channel) float64 24B 10.9 6.81 6.58
beamwidth_athwartship (channel) float64 24B 10.82 6.85 6.52
source_filenames (filenames) <U26 104B 'SOURCE FILE NOT IDE...
water_level float64 8B 9.15
Attributes:
processing_software_name: echopype
processing_software_version: 0.8.4
processing_time: 2024-04-25T21:30:48Z
processing_function: calibrate.compute_SvMVBS_ds
<xarray.Dataset> Size: 1MB
Dimensions: (channel: 3, ping_time: 391, echo_range: 150)
Coordinates:
* ping_time (ping_time) datetime64[ns] 3kB 2017-07-28T18:16:00 ......
* channel (channel) <U37 444B 'GPT 18 kHz 009072058c8d 1-1 ES18...
* echo_range (echo_range) float64 1kB 0.0 5.0 10.0 ... 740.0 745.0
Data variables:
Sv (channel, ping_time, echo_range) float64 1MB -15.13 .....
water_level float64 8B 9.15
frequency_nominal (channel) float64 24B 1.8e+04 3.8e+04 1.2e+05
Attributes:
processing_software_name: echopype
processing_software_version: 0.8.4
processing_time: 2024-04-25T21:30:50Z
processing_function: commongrid.compute_MVBSReplace the channel dimension and coordinate with the frequency_nominal variable containing actual frequency values. Note that this step is possible only because there are no duplicated frequencies present.
MVBS_ds = ep.consolidate.swap_dims_channel_frequency(MVBS_ds)
List the three frequencies used by the echosounder
MVBS_ds.frequency_nominal.values
array([ 18000., 38000., 120000.])
Generate an interactive plot of the three \(\text{MVBS}\) echograms, one for each frequency. The frequency can be changed via the slider widget.
import panel as pn
mvplot = MVBS_ds["Sv"].hvplot.image(
x='ping_time', y='echo_range',
color='Sv', rasterize=True,
cmap='jet', clim=(-80, -50),
xlabel='Time (UTC)',
ylabel='Depth (m)'
).options(width=500, invert_yaxis=True)
pn.panel(mvplot, center=True, widget_layout=pn.Row, widget_location='top_left')
1.6. Package versions#
import datetime
import numba, numpy, datashader, holoviews, s3fs, aiobotocore, botocore
print(f"echopype: {ep.__version__}")
print(
f"numpy: {numpy.__version__}, numba: {numba.__version__}, xarray: {xr.__version__}, geopandas: {gpd.__version__}\n"
f"fsspec: {fsspec.__version__}, s3fs: {s3fs.__version__}, aiobotocore: {aiobotocore.__version__}, botocore: {botocore.__version__}\n"
f"datashader: {datashader.__version__}, holoviews: {holoviews.__version__}, hvplot: {hvplot.__version__}"
)
print(f"\n{datetime.datetime.utcnow()} +00:00")
echopype: 0.8.4
numpy: 1.26.4, numba: 0.59.1, xarray: 2024.3.0, geopandas: 0.14.3
fsspec: 2024.3.1, s3fs: 2024.3.1, aiobotocore: 2.12.2, botocore: 1.34.51
datashader: 0.16.1, holoviews: 1.18.3, hvplot: 0.9.2
2024-04-25 21:30:51.605884 +00:00

